VLA Holography Tutorial¶
Important External Information¶
xarray Official Documentation (docs).
Dask Official Documentation (docs).
zarr Official Documentation (docs)
[ ]:
import os
try:
import astrohack
print('AstroHACK version', astrohack.__version__, 'already installed.')
except ImportError as e:
print(e)
print('Installing AstroHACK')
os.system("python -m pip install --upgrade astrohack")
import astrohack
print('astrohack version', astrohack.__version__, ' installed.')
Download Tutorial Data¶
[1]:
import astrohack
import graphviper
graphviper.utils.data.download('ea25_cal_small_after_fixed.split.ms', folder='data')
Holography Data File API¶
As part of the astroHACK API a set of functions to allow users to easily open on disk holography files has been provided. Each function takes an astroHACK holography file name as an argument and returns an object related to the given file type, ie. holog, image, panel, point. Each object allows the user to access data via dictionary keys with values consisting of the relevant holography dataset. Each object also provides a summary() helper function to list available keys for each
file. An example call for each file type is show below and the API documentation for all data-io functions can be found here.
from astrohack.dio import open_holog
from astrohack.dio import open_image
from astrohack.dio import open_panel
from astrohack.dio import open_pointing
holog_data = open_holog(file='./data/ea25_cal_small_spw1_4_60_ea04_after.holog.zarr')
image_data = open_image(file='./data/ea25_cal_small_spw1_4_60_ea04_after.image.zarr')
panel_data = open_panel(file='./data/ea25_cal_small_spw1_4_60_ea04_after.panel.zarr')
pointing_data = open_pointing(file='./data/ea25_cal_small_spw1_4_60_ea04_after.point.zarr')
Setup Dask Local Cluster¶
The local Dask client handles scheduling and worker managment for the parallelization. The user has the option of choosing the number of cores and memory allocations for each worker howerver, we recommend a minimum of 8Gb per core with standard settings.
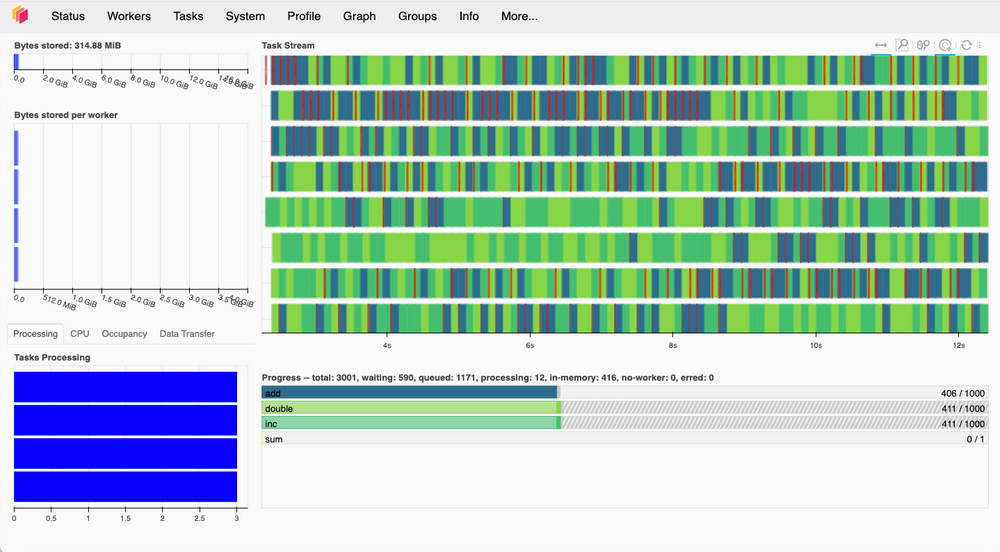
A significant amount of information related to the client and scheduling can be found using the Dask Dashboard. This is a built in dashboard native to Dask and allows the user to monitor the workers during processing. This is especially useful for profilling. For those that are interested in working soley within Jupyterlab a dashboard extension is availabe for Jupyterlab.
[2]:
from graphviper.dask.client import local_client
client = local_client(cores=2, memory_limit='8GB')
[2024-02-14 12:52:40,103] INFO graphviper: Checking parameter values for client.local_client
[2024-02-14 12:52:40,104] INFO logger: /export/home/ajax/jhoskins/.conda/envs/viper-3.10/lib/python3.10//site-packages/
[2024-02-14 12:52:40,104] INFO graphviper: Searching /export/home/ajax/jhoskins/.conda/envs/viper-3.10/lib/python3.10/site-packages/graphviper/config/ for configuration file, please wait ...
[2024-02-14 12:52:40,113] WARNING graphviper: It is recommended that the local cache directory be set using the dask_local_dir parameter.
[2024-02-14 12:52:41,487] INFO graphviper: Created client <MenrvaClient: 'tcp://127.0.0.1:33735' processes=2 threads=2, memory=14.90 GiB>
Extract Holog¶
The extraction and restructuring of the holography data is done using the extract_holog function. This function is similar in function to the UVHOL task in AIPS. The holography data that is extracted can be set using the compound dictionary holog_obs_description: mapping, scan, and antenna id. A detailed description of the structure of the holog_obs_description dictionary can be found in the documentation
here. The extract_holog can automatically generate the holog_obs_description by inspecting the pointing table.
Inline information on the input parameters can also be gotten using help(extract_holog) in the cell.
[3]:
from astrohack.extract_pointing import extract_pointing
from astrohack.extract_holog import extract_holog
extract_pointing(
ms_name='data/ea25_cal_small_after_fixed.split.ms',
point_name='data/ea25_cal_small_after_fixed.split.point.zarr',
parallel=True,
overwrite=True
)
holog_mds = extract_holog(
ms_name='data/ea25_cal_small_after_fixed.split.ms',
point_name='data/ea25_cal_small_after_fixed.split.point.zarr',
data_column='CORRECTED_DATA',
parallel=True,
overwrite=True
)
[2024-02-14 12:52:41,493] INFO graphviper: Checking parameter values for extract_pointing.extract_pointing
[2024-02-14 12:52:41,493] INFO astrohack: /export/home/ajax/jhoskins/Development/astrohack/
[2024-02-14 12:52:41,494] INFO graphviper: Searching /export/home/ajax/jhoskins/.conda/envs/viper-3.10/lib/python3.10/site-packages/graphviper/config/ for configuration file, please wait ...
[2024-02-14 12:52:41,495] INFO astrohack: Failed to find module in PARAMETER_CONFIG_PATH ... attempting to check common directories ...
[2024-02-14 12:52:41,495] INFO graphviper: Searching /export/home/ajax/jhoskins/Development/astrohack/ for configuration file, please wait ...
Successful readonly open of usernoread-locked table data/ea25_cal_small_after_fixed.split.ms/ANTENNA: 8 columns, 3 rows
[2024-02-14 12:52:47,678] INFO graphviper: Finished processing
[2024-02-14 12:52:47,712] INFO graphviper: Checking parameter values for extract_holog.extract_holog
[2024-02-14 12:52:47,713] INFO astrohack: /export/home/ajax/jhoskins/Development/astrohack/
[2024-02-14 12:52:47,713] INFO graphviper: Searching /export/home/ajax/jhoskins/.conda/envs/viper-3.10/lib/python3.10/site-packages/graphviper/config/ for configuration file, please wait ...
[2024-02-14 12:52:47,714] INFO astrohack: Failed to find module in PARAMETER_CONFIG_PATH ... attempting to check common directories ...
[2024-02-14 12:52:47,714] INFO graphviper: Searching /export/home/ajax/jhoskins/Development/astrohack/ for configuration file, please wait ...
[2024-02-14 12:52:47,760] INFO graphviper: Creating output file name: data/ea25_cal_small_after_fixed.split.holog.zarr
Successful readonly open of usernoread-locked table data/ea25_cal_small_after_fixed.split.ms/DATA_DESCRIPTION: 3 columns, 2 rows
Successful readonly open of usernoread-locked table data/ea25_cal_small_after_fixed.split.ms/ANTENNA: 8 columns, 3 rows
Successful readonly open of usernoread-locked table data/ea25_cal_small_after_fixed.split.ms: 23 columns, 36580 rows
Successful readonly open of usernoread-locked table data/ea25_cal_small_after_fixed.split.ms/STATE: 7 columns, 44 rows
Successful readonly open of usernoread-locked table data/ea25_cal_small_after_fixed.split.ms/SPECTRAL_WINDOW: 20 columns, 2 rows
Successful readonly open of usernoread-locked table data/ea25_cal_small_after_fixed.split.ms/POLARIZATION: 4 columns, 1 rows
Successful readonly open of usernoread-locked table data/ea25_cal_small_after_fixed.split.ms/OBSERVATION: 9 columns, 1 rows
Successful readonly open of usernoread-locked table data/ea25_cal_small_after_fixed.split.ms/HISTORY: 9 columns, 157 rows
[2024-02-14 12:52:47,804] INFO graphviper: Processing ddi: 0, scans: [8 ... 57]
[2024-02-14 12:52:47,805] INFO graphviper: Processing ddi: 1, scans: [8 ... 57]
Successful readonly open of default-locked table data/ea25_cal_small_after_fixed.split.ms/ANTENNA: 8 columns, 3 rows
Successful readonly open of default-locked table data/ea25_cal_small_after_fixed.split.ms/OBSERVATION: 9 columns, 1 rows
[2024-02-14 12:53:01,368] INFO worker_0: Writing holog file to data/ea25_cal_small_after_fixed.split.holog.zarr
[2024-02-14 12:53:01,542] INFO worker_0: Writing holog file to data/ea25_cal_small_after_fixed.split.holog.zarr
Successful readonly open of default-locked table data/ea25_cal_small_after_fixed.split.ms/ANTENNA: 8 columns, 3 rows
Successful readonly open of default-locked table data/ea25_cal_small_after_fixed.split.ms/OBSERVATION: 9 columns, 1 rows
[2024-02-14 12:53:01,648] INFO worker_1: Writing holog file to data/ea25_cal_small_after_fixed.split.holog.zarr
[2024-02-14 12:53:01,663] INFO worker_0: Finished extracting holography chunk for ddi: 1 holog_map_key: map_0
[2024-02-14 12:53:01,854] INFO worker_1: Writing holog file to data/ea25_cal_small_after_fixed.split.holog.zarr
[2024-02-14 12:53:02,079] INFO worker_1: Finished extracting holography chunk for ddi: 0 holog_map_key: map_0
[2024-02-14 12:53:02,094] INFO graphviper: Finished processing
[2024-02-14 12:53:02,136] WARNING graphviper: Cell size not consistent: [0.000719484970141879, 0.000719484970141879, 0.0006386556122807017, 0.0006386556122807017]
[2024-02-14 12:53:02,137] WARNING graphviper: Calculating suggested cell size ...
[2024-02-14 12:53:02,138] WARNING graphviper: [0.00071948 0.00071948 0.00063866 0.00063866]
[2024-02-14 12:53:02,141] INFO graphviper: The suggested cell size is calculated to be: 0.0006790702912112905
[2024-02-14 12:53:02,144] WARNING graphviper: Number of pixels not consistent: [729, 729, 961, 961]
[2024-02-14 12:53:02,147] WARNING graphviper: Calculating suggested number of pixels ...
[2024-02-14 12:53:02,148] WARNING graphviper: [729 729 961 961]
[2024-02-14 12:53:02,150] INFO graphviper: The suggested number of pixels is calculated to be: 845 (grid: 29 x 29)
Two files are created by extract_holog: The extracted pointing information in the form of <point_name>.point.zarr and the extracted holography data as <point_name>.holog.zarr. In addition, a holography data object is returned. This is the same holography data object returned by the hologrphy data API above. The holog_mds object is a python dict containing the extracted holography data found in .holog.zarr but with extended functionality such as providing a summary of the run
infomation in table form. Below for each DDI we can see the available scan and antenna information.
point_name.point.zarr: The pointing zarr file contains position and pointing information extracted from the pointing table of the input measurement set. In addition, the antenna and mapping scan information is listed for each antenna. The pointing object is structured as a simple dictionary with key:value sets with the key being the antenna id and the value being the pointing dataset.
point_mds =
{
ant_0: point_ds,
⋮
ant_n: point_ds
}
holog_name.holog.zarr: The holog zarr file contains ungridded data extracted from the pointing and main tables in the measurement set. The holog file includes the directional, visibility and weight information recorded on a shared time axis; the sampling is done because the native sample rates between the pointing and main tables are not the same. In addition, the meta data such as sampled parallactic data (beginning, middle and end of scan) and l(m) extent is recorded in the file
attributes. The holog file structure is a compound dictionary keyed according to ddi -> map -> ant with values consisting of the holog dataset.
holog_mds =
{
ddi_0:{
map_0:{
ant_0: holog_ds,
⋮
ant_n: holog_ds
},
⋮
map_p: …
},
⋮
ddi_m: …
}
An example of the holog dataset object is show below.
[4]:
holog_mds['ddi_0']['map_0']['ant_ea25']
[4]:
<xarray.Dataset>
Dimensions: (time: 8914, lm: 2, chan: 64, pol: 4)
Coordinates:
* chan (chan) float64 1.41e+10 1.411e+10 ... 1.423e+10
* pol (pol) <U2 'RR' 'RL' 'LR' 'LL'
* time (time) float64 5.17e+09 5.17e+09 ... 5.17e+09
Dimensions without coordinates: lm
Data variables:
DIRECTIONAL_COSINES (time, lm) float64 dask.array<chunksize=(8914, 2), meta=np.ndarray>
IDEAL_DIRECTIONAL_COSINES (time, lm) float64 dask.array<chunksize=(8914, 2), meta=np.ndarray>
VIS (time, chan, pol) complex64 dask.array<chunksize=(2229, 16, 2), meta=np.ndarray>
WEIGHT (time, chan, pol) float64 dask.array<chunksize=(2229, 16, 2), meta=np.ndarray>
Attributes:
antenna_name: ea25
ddi: 0
grid_params: {'cell_size': 0.000719484970141879, 'n_pix': 729}
holog_map_key: map_0
l_max: 0.009675024871632517
l_min: -0.009674930411087166
m_max: 0.009698666161358812
m_min: -0.00975229251000802
parallactic_samples: [5.308157433326323, 5.357028871639436, 5.502977...
telescope_name: EVLA
time_smoothing_interval: 1.0In this case, there is only one selection in the holography file as seen in the summary. Using the available keys we can see an overview of the Dask dataset structure. In addition, the numpy arrays for the data are accessed by calling values on a given dataset variable. For instance accessing the data for the DIRECTIONAL_COSINES below would be simply
>> holog_mds['ddi_0']['map_0']['ant_ea25'].DIRECTIONAL_COSINES.values
>> array([[-0.00433549, -0.0027946 ],
[-0.00870191, -0.00682571],
[-0.00965634, -0.00908509],
...,
[ 0.00966373, 0.00957556],
[ 0.00966267, 0.00957601],
[ 0.00965895, 0.00956941]])
>> holog_mds['ddi_0']['map_0']['ant_ea25'].DIRECTIONAL_COSINES.values.shape
>> (9145, 2)
where the dimension are given in the mds output for each data variable (in this case (time, lm)). A more in-depth overview of how to interact with Dask dataset can be found here.
A summary of the available key values can be obtained using the summary convenience function.
[5]:
holog_mds.summary()
####################################################################################################
### Summary for: ###
### data/ea25_cal_small_after_fixed.split.holog.zarr ###
####################################################################################################
Full documentation for AstrohackHologFile objects' API at:
https://astrohack.readthedocs.io/en/stable/_api/autoapi/astrohack/mds/index.html#astrohack.mds.AstrohackHologFile
Input Parameters:
+---------------------------+--------------------------------------------------+
| Parameter | Value |
+---------------------------+--------------------------------------------------+
| ms_name | data/ea25_cal_small_after_fixed.split.ms |
| point_name | data/ea25_cal_small_after_fixed.split.point.zarr |
| holog_name | data/ea25_cal_small_after_fixed.split.holog.zarr |
| holog_obs_dict | None |
| ddi | all |
| baseline_average_distance | all |
| baseline_average_nearest | all |
| data_column | CORRECTED_DATA |
| time_smoothing_interval | None |
| parallel | True |
| overwrite | True |
| version | 0.4.5 |
| origin | extract_holog |
+---------------------------+--------------------------------------------------+
Contents:
+-------+-------+--------------------------+
| DDI | Map | Antenna |
+-------+-------+--------------------------+
| ddi_0 | map_0 | ['ant_ea25', 'ant_ea06'] |
| ddi_1 | map_0 | ['ant_ea25', 'ant_ea06'] |
+-------+-------+--------------------------+
Available methods:
+----------------------+-----------------------------------------------------------------------+
| Methods | Description |
+----------------------+-----------------------------------------------------------------------+
| summary | Prints summary of the AstrohackHologFile object, with available data, |
| | attributes and available methods |
| select | Select data on the basis of ddi, scan, ant. This is a convenience |
| | function. |
| plot_diagnostics | Plot diagnostic calibration plots from the holography data file. |
| plot_lm_sky_coverage | Plot directional cosine coverage. |
+----------------------+-----------------------------------------------------------------------+
Estimated Memory Requirements¶
A new functionality, currently being refined, is a function to estimate the amount of memory per core max that would be required to process a given file. The estimation is given as the suggested memory per core need to not spilling over into swap memory. If the user has already computed the holog_obs_dict, it can be added as a parameter to speed up the estitmate as this is a serial function currently.
In the resulting table the following definitions are important:
Available memory: The available memory on the system currently, ie. the total not currently in use.
Total memory: The total system memory
Suggested memory per core: Memory allocation per core estimated to not spill ove rinto swap memory.
Reference: https://psutil.readthedocs.io/en/latest/#psutil.virtual_memory
[6]:
from astrohack.extract_holog import model_memory_usage
from astrohack.extract_holog import generate_holog_obs_dict
holog_obs_dict = generate_holog_obs_dict(
ms_name="data/ea25_cal_small_after_fixed.split.ms",
point_name="data/ea25_cal_small_after_fixed.split.point.zarr",
)
model_memory_usage(
ms_name="data/ea25_cal_small_after_fixed.split.ms",
holog_obs_dict=holog_obs_dict
)
Successful readonly open of usernoread-locked table data/ea25_cal_small_after_fixed.split.ms/ANTENNA: 8 columns, 3 rows
Successful readonly open of usernoread-locked table data/ea25_cal_small_after_fixed.split.ms: 23 columns, 36580 rows
[2024-02-14 12:53:02,344] INFO graphviper: Writing distance matrix to /export/home/ajax/jhoskins/Development/astrohack/docs/.baseline_distance_matrix.csv ...
/export/home/ajax/jhoskins/.conda/envs/viper-3.10/lib/python3.10/site-packages/sklearn/base.py:376: InconsistentVersionWarning: Trying to unpickle estimator ElasticNet from version 1.3.2 when using version 1.4.0. This might lead to breaking code or invalid results. Use at your own risk. For more info please refer to:
https://scikit-learn.org/stable/model_persistence.html#security-maintainability-limitations
warnings.warn(
System Info ┏━━━━━━━━━┳━━━━━━━━━━━━━━━━━━━━━━━┳━━━━━━━━━━━━━━━━━━━┳━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━┓ ┃ N-cores ┃ Available memory (MB) ┃ Total memory (MB) ┃ Suggested memory per core (MB) ┃ ┡━━━━━━━━━╇━━━━━━━━━━━━━━━━━━━━━━━╇━━━━━━━━━━━━━━━━━━━╇━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━┩ │ 4 │ 3017 │ 15881 │ 11911 │ └─────────┴───────────────────────┴───────────────────┴────────────────────────────────┘ Available memory: represents the system memory available without going into swap
[6]:
11911
Holog¶
The holog function processes the holography data and produces a holog image file on disk with the suffix, .image.zarr. This function is a direct replacement for the task HOLOG in AIPS. It is required that the user provide the grid_size and cell_size when processing holography data. The grid_size defines the number of l x m points used to when doing the gridding. The cell_size defines the value in arseconds of each grid spacing. More in-depth parameter information
can be found in readthedocs here.
Inline information on the input paramters can also be gotten using help(holog) in the cell.
[7]:
import numpy as np
from astrohack import holog
cell_size = np.array([-0.0006442, 0.0006442]) # arcseconds
grid_size = np.array([31, 31]) # pixels
image_mds = holog(
holog_name='data/ea25_cal_small_after_fixed.split.holog.zarr',
overwrite=True,
phase_fit=True,
apply_mask=True,
to_stokes=True,
parallel=True
)
[2024-02-14 12:53:09,721] INFO graphviper: Checking parameter values for holog.holog
[2024-02-14 12:53:09,722] INFO astrohack: /export/home/ajax/jhoskins/Development/astrohack/
[2024-02-14 12:53:09,723] INFO graphviper: Searching /export/home/ajax/jhoskins/.conda/envs/viper-3.10/lib/python3.10/site-packages/graphviper/config/ for configuration file, please wait ...
[2024-02-14 12:53:09,725] INFO astrohack: Failed to find module in PARAMETER_CONFIG_PATH ... attempting to check common directories ...
[2024-02-14 12:53:09,726] INFO graphviper: Searching /export/home/ajax/jhoskins/Development/astrohack/ for configuration file, please wait ...
[2024-02-14 12:53:09,772] INFO graphviper: Creating output file name: data/ea25_cal_small_after_fixed.split.image.zarr
[2024-02-14 12:53:09,774] INFO graphviper: Cell size: [-0.00067907 0.00067907], Grid size [29 29]
/export/home/ajax/jhoskins/Development/astrohack/src/astrohack/_utils/_holog.py:68: FutureWarning: The return type of `Dataset.dims` will be changed to return a set of dimension names in future, in order to be more consistent with `DataArray.dims`. To access a mapping from dimension names to lengths, please use `Dataset.sizes`.
n_chan = ant_data_dict[ddi][map0].dims["chan"]
/export/home/ajax/jhoskins/Development/astrohack/src/astrohack/_utils/_holog.py:69: FutureWarning: The return type of `Dataset.dims` will be changed to return a set of dimension names in future, in order to be more consistent with `DataArray.dims`. To access a mapping from dimension names to lengths, please use `Dataset.sizes`.
n_pol = ant_data_dict[ddi][map0].dims["pol"]
/export/home/ajax/jhoskins/Development/astrohack/src/astrohack/_utils/_holog.py:68: FutureWarning: The return type of `Dataset.dims` will be changed to return a set of dimension names in future, in order to be more consistent with `DataArray.dims`. To access a mapping from dimension names to lengths, please use `Dataset.sizes`.
n_chan = ant_data_dict[ddi][map0].dims["chan"]
/export/home/ajax/jhoskins/Development/astrohack/src/astrohack/_utils/_holog.py:69: FutureWarning: The return type of `Dataset.dims` will be changed to return a set of dimension names in future, in order to be more consistent with `DataArray.dims`. To access a mapping from dimension names to lengths, please use `Dataset.sizes`.
n_pol = ant_data_dict[ddi][map0].dims["pol"]
/export/home/ajax/jhoskins/Development/astrohack/src/astrohack/_utils/_holog.py:119: FutureWarning: The return type of `Dataset.dims` will be changed to return a set of dimension names in future, in order to be more consistent with `DataArray.dims`. To access a mapping from dimension names to lengths, please use `Dataset.sizes`.
time_centroid_index = ant_data_dict[ddi][holog_map].dims["time"] // 2
/export/home/ajax/jhoskins/Development/astrohack/src/astrohack/_utils/_holog.py:119: FutureWarning: The return type of `Dataset.dims` will be changed to return a set of dimension names in future, in order to be more consistent with `DataArray.dims`. To access a mapping from dimension names to lengths, please use `Dataset.sizes`.
time_centroid_index = ant_data_dict[ddi][holog_map].dims["time"] // 2
[2024-02-14 12:53:10,180] INFO worker_1: Calculating aperture pattern ...
[2024-02-14 12:53:10,180] INFO worker_1: Calculating aperture illumination pattern ...
[2024-02-14 12:53:10,181] INFO worker_0: Calculating aperture pattern ...
[2024-02-14 12:53:10,181] INFO worker_0: Calculating aperture illumination pattern ...
[2024-02-14 12:53:10,502] INFO worker_0: Applying phase correction
[2024-02-14 12:53:10,554] INFO worker_1: Applying phase correction
/export/home/ajax/jhoskins/Development/astrohack/src/astrohack/_utils/_holog.py:68: FutureWarning: The return type of `Dataset.dims` will be changed to return a set of dimension names in future, in order to be more consistent with `DataArray.dims`. To access a mapping from dimension names to lengths, please use `Dataset.sizes`.
n_chan = ant_data_dict[ddi][map0].dims["chan"]
/export/home/ajax/jhoskins/Development/astrohack/src/astrohack/_utils/_holog.py:69: FutureWarning: The return type of `Dataset.dims` will be changed to return a set of dimension names in future, in order to be more consistent with `DataArray.dims`. To access a mapping from dimension names to lengths, please use `Dataset.sizes`.
n_pol = ant_data_dict[ddi][map0].dims["pol"]
/export/home/ajax/jhoskins/Development/astrohack/src/astrohack/_utils/_holog.py:68: FutureWarning: The return type of `Dataset.dims` will be changed to return a set of dimension names in future, in order to be more consistent with `DataArray.dims`. To access a mapping from dimension names to lengths, please use `Dataset.sizes`.
n_chan = ant_data_dict[ddi][map0].dims["chan"]
/export/home/ajax/jhoskins/Development/astrohack/src/astrohack/_utils/_holog.py:69: FutureWarning: The return type of `Dataset.dims` will be changed to return a set of dimension names in future, in order to be more consistent with `DataArray.dims`. To access a mapping from dimension names to lengths, please use `Dataset.sizes`.
n_pol = ant_data_dict[ddi][map0].dims["pol"]
/export/home/ajax/jhoskins/Development/astrohack/src/astrohack/_utils/_holog.py:119: FutureWarning: The return type of `Dataset.dims` will be changed to return a set of dimension names in future, in order to be more consistent with `DataArray.dims`. To access a mapping from dimension names to lengths, please use `Dataset.sizes`.
time_centroid_index = ant_data_dict[ddi][holog_map].dims["time"] // 2
[2024-02-14 12:53:15,146] INFO worker_0: Calculating aperture pattern ...
[2024-02-14 12:53:15,146] INFO worker_0: Calculating aperture illumination pattern ...
/export/home/ajax/jhoskins/Development/astrohack/src/astrohack/_utils/_holog.py:119: FutureWarning: The return type of `Dataset.dims` will be changed to return a set of dimension names in future, in order to be more consistent with `DataArray.dims`. To access a mapping from dimension names to lengths, please use `Dataset.sizes`.
time_centroid_index = ant_data_dict[ddi][holog_map].dims["time"] // 2
[2024-02-14 12:53:15,411] INFO worker_1: Calculating aperture pattern ...
[2024-02-14 12:53:15,411] INFO worker_1: Calculating aperture illumination pattern ...
[2024-02-14 12:53:15,462] INFO worker_0: Applying phase correction
[2024-02-14 12:53:15,609] INFO worker_1: Applying phase correction
[2024-02-14 12:53:15,873] INFO graphviper: Finished processing
image_name.image.zarr: The image zarr file contains gridded image data the beam, extracted aperture and the amplitude and phase components. It also contains all of the relevant coordinate information. The image file structure is a compound dictionary keys according to ant -> ddi with the dictionary values consisting of the image dataset.
image_mds =
{
ant_0:{
ddi_0: image_ds,
⋮
ddi_m: image_ds
},
⋮
ant_n: …
}
An example of the image dataset object is show below.
[8]:
image_mds['ant_ea25']['ddi_0']
[8]:
<xarray.Dataset>
Dimensions: (time: 1, chan: 1, pol: 4, u_prime: 552, v_prime: 552,
u: 627, v: 627, l: 29, m: 29, ddi: 1)
Coordinates:
* chan (chan) float64 1.417e+10
* ddi (ddi) <U5 'ddi_0'
* l (l) float64 0.009507 0.008828 ... -0.008828 -0.009507
* m (m) float64 -0.009507 -0.008828 ... 0.008828 0.009507
* pol (pol) <U1 'I' 'Q' 'U' 'V'
* u (u) float64 735.1 732.8 730.4 ... -730.4 -732.8 -735.1
* u_prime (u_prime) float64 648.2 645.9 643.5 ... -643.5 -645.9
* v (v) float64 -735.1 -732.8 -730.4 ... 730.4 732.8 735.1
* v_prime (v_prime) float64 -648.2 -645.9 -643.5 ... 643.5 645.9
Dimensions without coordinates: time
Data variables:
AMPLITUDE (time, chan, pol, u_prime, v_prime) float64 dask.array<chunksize=(1, 1, 1, 276, 276), meta=np.ndarray>
APERTURE (time, chan, pol, u, v) complex128 dask.array<chunksize=(1, 1, 1, 157, 314), meta=np.ndarray>
BEAM (time, chan, pol, l, m) complex128 dask.array<chunksize=(1, 1, 4, 29, 29), meta=np.ndarray>
CORRECTED_PHASE (time, chan, pol, u_prime, v_prime) float64 dask.array<chunksize=(1, 1, 1, 276, 276), meta=np.ndarray>
Attributes:
ant_id: ant_ea25
ant_name: ea25
aperture_resolution: [1.413428251773375, 1.413428251773375]
ddi: ddi_0
telescope_name: EVLA
time_centroid: 5170359434.000001A summary of the available key values can be obtained using the summary convenience function.
[9]:
image_mds.summary()
####################################################################################################
### Summary for: ###
### data/ea25_cal_small_after_fixed.split.image.zarr ###
####################################################################################################
Full documentation for AstrohackImageFile objects' API at:
https://astrohack.readthedocs.io/en/stable/_api/autoapi/astrohack/mds/index.html#astrohack.mds.AstrohackImageFile
Input Parameters:
+-------------------------+--------------------------------------------------+
| Parameter | Value |
+-------------------------+--------------------------------------------------+
| holog_name | data/ea25_cal_small_after_fixed.split.holog.zarr |
| grid_size | None |
| cell_size | None |
| image_name | data/ea25_cal_small_after_fixed.split.image.zarr |
| padding_factor | 50 |
| grid_interpolation_mode | linear |
| chan_average | True |
| chan_tolerance_factor | 0.005 |
| scan_average | True |
| ant | all |
| ddi | all |
| to_stokes | True |
| apply_mask | True |
| phase_fit | True |
| overwrite | True |
| parallel | True |
| version | 0.4.5 |
| origin | holog |
+-------------------------+--------------------------------------------------+
Contents:
+----------+--------------------+
| Antenna | DDI |
+----------+--------------------+
| ant_ea25 | ['ddi_0', 'ddi_1'] |
| ant_ea06 | ['ddi_0', 'ddi_1'] |
+----------+--------------------+
Available methods:
+----------------+------------------------------------------------------------------------------+
| Methods | Description |
+----------------+------------------------------------------------------------------------------+
| summary | Prints summary of the AstrohackImageFile object, with available data, |
| | attributes and available methods |
| select | Select data on the basis of ddi, scan, ant. This is a convenience function. |
| export_to_fits | Export contents of an AstrohackImageFile object to several FITS files in the |
| | destination folder |
| plot_beams | Beam plots from the data in an AstrohackImageFIle object. |
| plot_apertures | Aperture amplitude and phase plots from the data in an AstrohackImageFIle |
| | object. |
+----------------+------------------------------------------------------------------------------+
Each of the holography output files is a compound dictionary with respect to the run parameters and contains a xarray Dataset, this means that the holography files have access to all native xarray functionality. The user can use their favorite plotting package to visualize the data or use xarray’s internal functions to do simple filtering and plotting.
[10]:
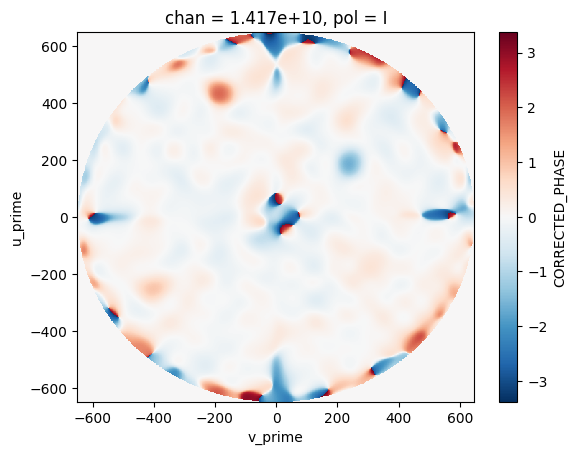
image_mds['ant_ea25']['ddi_0'].CORRECTED_PHASE.isel(chan=0, pol=0).plot()
[10]:
<matplotlib.collections.QuadMesh at 0x7f18f7d3e620>
Panel¶
The panel function takes the place of and expands the PANEL AIPS function to processes the image information and derives adjustements to the dish panels. This produces a file on disk of format .panel.zarr containing information on corrections, residuals and screw adjustments. As an added bonus the panel function has a helper function to convert aips data to astrohack format and process it using the aips_holog_to_astrohack function. For a full description of the operation and
arguments of the panel function see docs.
[11]:
from astrohack.panel import panel
panel_model = 'rigid'
panel_mds = panel(
image_name='data/ea25_cal_small_after_fixed.split.image.zarr',
panel_model=panel_model,
panel_margins=0.2,
clip_type='relative',
clip_level=0.2,
parallel=True,
overwrite=True
)
[2024-02-14 12:53:36,343] INFO graphviper: Checking parameter values for panel.panel
[2024-02-14 12:53:36,345] INFO astrohack: /export/home/ajax/jhoskins/Development/astrohack/
[2024-02-14 12:53:36,346] INFO graphviper: Searching /export/home/ajax/jhoskins/.conda/envs/viper-3.10/lib/python3.10/site-packages/graphviper/config/ for configuration file, please wait ...
[2024-02-14 12:53:36,347] INFO astrohack: Failed to find module in PARAMETER_CONFIG_PATH ... attempting to check common directories ...
[2024-02-14 12:53:36,347] INFO graphviper: Searching /export/home/ajax/jhoskins/Development/astrohack/ for configuration file, please wait ...
[2024-02-14 12:53:36,395] INFO graphviper: Creating output file name: data/ea25_cal_small_after_fixed.split.panel.zarr
[2024-02-14 12:53:37,189] INFO worker_0: processing ant_ea25 ddi_0
/export/home/ajax/jhoskins/Development/astrohack/src/astrohack/_utils/_panel_classes/antenna_surface.py:77: FutureWarning: The return type of `Dataset.dims` will be changed to return a set of dimension names in future, in order to be more consistent with `DataArray.dims`. To access a mapping from dimension names to lengths, please use `Dataset.sizes`.
if inputxds.dims['chan'] != 1:
/export/home/ajax/jhoskins/Development/astrohack/src/astrohack/_utils/_panel_classes/antenna_surface.py:93: FutureWarning: The return type of `Dataset.dims` will be changed to return a set of dimension names in future, in order to be more consistent with `DataArray.dims`. To access a mapping from dimension names to lengths, please use `Dataset.sizes`.
self.npoint = np.sqrt(inputxds.dims['l'] ** 2 + inputxds.dims['m'] ** 2)
[2024-02-14 12:53:37,529] INFO worker_1: processing ant_ea25 ddi_1
/export/home/ajax/jhoskins/Development/astrohack/src/astrohack/_utils/_panel_classes/antenna_surface.py:77: FutureWarning: The return type of `Dataset.dims` will be changed to return a set of dimension names in future, in order to be more consistent with `DataArray.dims`. To access a mapping from dimension names to lengths, please use `Dataset.sizes`.
if inputxds.dims['chan'] != 1:
/export/home/ajax/jhoskins/Development/astrohack/src/astrohack/_utils/_panel_classes/antenna_surface.py:93: FutureWarning: The return type of `Dataset.dims` will be changed to return a set of dimension names in future, in order to be more consistent with `DataArray.dims`. To access a mapping from dimension names to lengths, please use `Dataset.sizes`.
self.npoint = np.sqrt(inputxds.dims['l'] ** 2 + inputxds.dims['m'] ** 2)
[2024-02-14 12:53:40,159] INFO worker_0: processing ant_ea06 ddi_0
/export/home/ajax/jhoskins/Development/astrohack/src/astrohack/_utils/_panel_classes/antenna_surface.py:77: FutureWarning: The return type of `Dataset.dims` will be changed to return a set of dimension names in future, in order to be more consistent with `DataArray.dims`. To access a mapping from dimension names to lengths, please use `Dataset.sizes`.
if inputxds.dims['chan'] != 1:
/export/home/ajax/jhoskins/Development/astrohack/src/astrohack/_utils/_panel_classes/antenna_surface.py:93: FutureWarning: The return type of `Dataset.dims` will be changed to return a set of dimension names in future, in order to be more consistent with `DataArray.dims`. To access a mapping from dimension names to lengths, please use `Dataset.sizes`.
self.npoint = np.sqrt(inputxds.dims['l'] ** 2 + inputxds.dims['m'] ** 2)
[2024-02-14 12:53:41,502] INFO worker_1: processing ant_ea06 ddi_1
/export/home/ajax/jhoskins/Development/astrohack/src/astrohack/_utils/_panel_classes/antenna_surface.py:77: FutureWarning: The return type of `Dataset.dims` will be changed to return a set of dimension names in future, in order to be more consistent with `DataArray.dims`. To access a mapping from dimension names to lengths, please use `Dataset.sizes`.
if inputxds.dims['chan'] != 1:
/export/home/ajax/jhoskins/Development/astrohack/src/astrohack/_utils/_panel_classes/antenna_surface.py:93: FutureWarning: The return type of `Dataset.dims` will be changed to return a set of dimension names in future, in order to be more consistent with `DataArray.dims`. To access a mapping from dimension names to lengths, please use `Dataset.sizes`.
self.npoint = np.sqrt(inputxds.dims['l'] ** 2 + inputxds.dims['m'] ** 2)
[2024-02-14 12:53:43,122] WARNING worker_1: Fit failed with the rigid model and a simple mean has been used instead for the following panels: ['ea06', 'ddi_1']
[2024-02-14 12:53:43,122] WARNING worker_1: ['6-10']
[2024-02-14 12:53:43,762] INFO graphviper: Finished processing
panel_name.panel.zarr: The panel zarr file contains process information regarding the per panel screw corrections as well as residuals, masks and phase corrections used to produce them. The panel file structure is a compound dictionary keyed according to ant -> ddi with the value consisting of the panel dataset.
panel_mds =
{
ant_0:{
ddi_0: panel_ds,
⋮
ddi_m: panel_ds
},
⋮
ant_n: …
An example of the panel dataset object is show below.
[12]:
panel_mds['ant_ea25']['ddi_0']
[12]:
<xarray.Dataset>
Dimensions: (u: 552, v: 552, labels: 172, pars: 3, screws: 4)
Coordinates:
* labels (labels) object '1-1' '1-2' '1-3' ... '6-39' '6-40'
* pars (pars) int64 0 1 2
* screws (screws) <U2 'il' 'ir' 'ol' 'or'
* u (u) float64 13.72 13.67 13.62 ... -13.57 -13.62 -13.67
* v (v) float64 -13.72 -13.67 -13.62 ... 13.57 13.62 13.67
Data variables:
AMPLITUDE (u, v) float64 dask.array<chunksize=(138, 276), meta=np.ndarray>
CORRECTIONS (u, v) float64 dask.array<chunksize=(138, 276), meta=np.ndarray>
DEVIATION (u, v) float64 dask.array<chunksize=(138, 276), meta=np.ndarray>
MASK (u, v) bool dask.array<chunksize=(276, 552), meta=np.ndarray>
PANEL_DISTRIBUTION (u, v) int32 dask.array<chunksize=(276, 276), meta=np.ndarray>
PANEL_PARAMETERS (labels, pars) float64 dask.array<chunksize=(172, 3), meta=np.ndarray>
PANEL_SCREWS (labels, screws) float64 dask.array<chunksize=(172, 4), meta=np.ndarray>
PHASE (u, v) float64 dask.array<chunksize=(138, 276), meta=np.ndarray>
PHASE_CORRECTIONS (u, v) float64 dask.array<chunksize=(138, 276), meta=np.ndarray>
PHASE_RESIDUALS (u, v) float64 dask.array<chunksize=(138, 276), meta=np.ndarray>
RESIDUALS (u, v) float64 dask.array<chunksize=(138, 276), meta=np.ndarray>
Attributes: (12/16)
amp_unit: V
ant_name: ea25
aperture_resolution: [1.413428251773375, 1.413428251773375]
clip: 0.7180989426650104
ddi: ddi_0
fitted: True
... ...
panel_margin: 0.2
panel_model: rigid
solved: True
telescope_name: VLA
theoretical_gain: 100.1817827185667
wavelength: 0.021161322651231735A summary of the available key values can be obtained using the summary convenience function.
[13]:
panel_mds.summary()
####################################################################################################
### Summary for: ###
### data/ea25_cal_small_after_fixed.split.panel.zarr ###
####################################################################################################
Full documentation for AstrohackPanelFile objects' API at:
https://astrohack.readthedocs.io/en/stable/_api/autoapi/astrohack/mds/index.html#astrohack.mds.AstrohackPanelFile
Input Parameters:
+--------------------+--------------------------------------------------+
| Parameter | Value |
+--------------------+--------------------------------------------------+
| image_name | data/ea25_cal_small_after_fixed.split.image.zarr |
| panel_name | data/ea25_cal_small_after_fixed.split.panel.zarr |
| clip_type | relative |
| clip_level | 0.2 |
| panel_model | rigid |
| panel_margins | 0.2 |
| polarization_state | I |
| ant | all |
| ddi | all |
| parallel | True |
| overwrite | True |
| version | 0.4.5 |
| origin | panel |
+--------------------+--------------------------------------------------+
Contents:
+----------+--------------------+
| Antenna | DDI |
+----------+--------------------+
| ant_ea25 | ['ddi_0', 'ddi_1'] |
| ant_ea06 | ['ddi_0', 'ddi_1'] |
+----------+--------------------+
Available methods:
+----------------+-----------------------------------------------------------------------+
| Methods | Description |
+----------------+-----------------------------------------------------------------------+
| summary | Prints summary of the AstrohackPanelFile object, with available data, |
| | attributes and available methods |
| get_antenna | Retrieve an AntennaSurface object for interaction |
| export_screws | Export screw adjustments to text files and optionally plots. |
| export_to_fits | Export contents of an Astrohack MDS file to several FITS files in the |
| | destination folder |
| plot_antennas | Create diagnostic plots of antenna surfaces from panel data file. |
+----------------+-----------------------------------------------------------------------+
Additional Functions¶
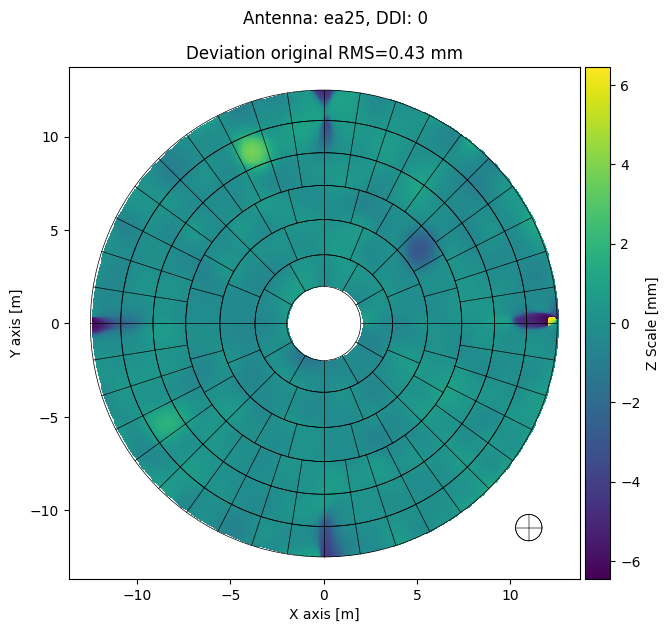
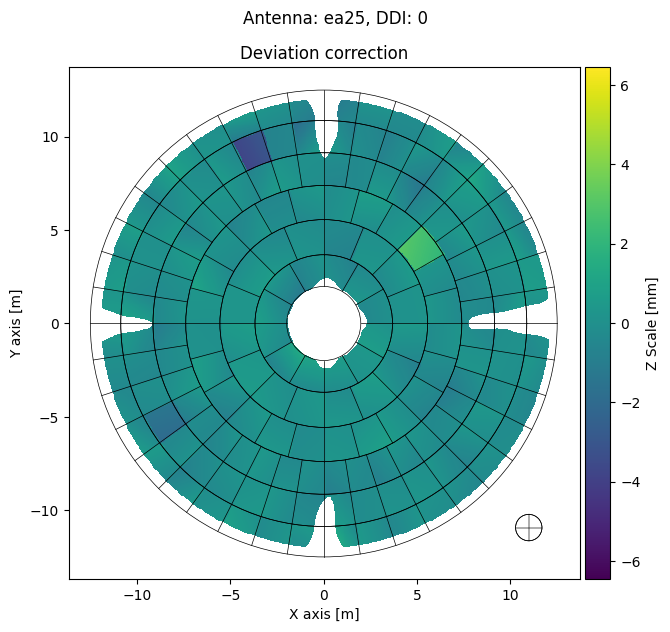
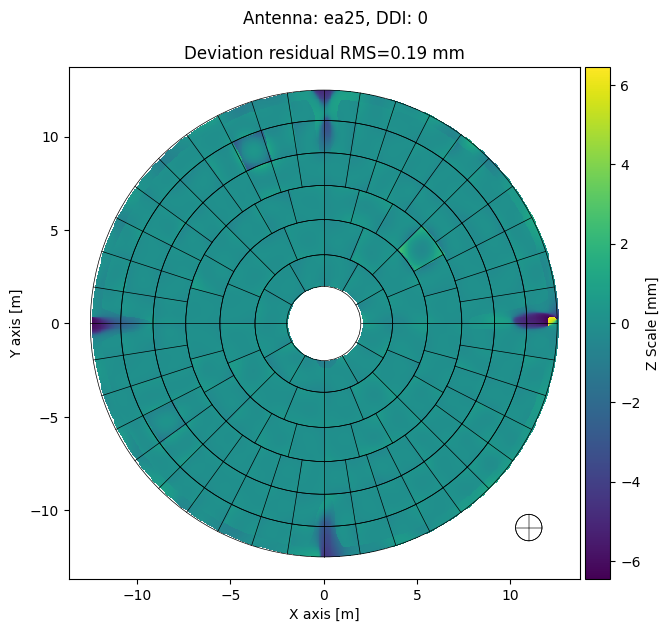
The panel_mds object provides two helper functions for the user to export or investigate the results of the panel function. - export_screws(): This method exports the screw and panel adjustements from the panel output file. - plot_antennas(): This method plots one of three diagnostics plots from the panel output file data. The plots types are: deviation, phase and ancillary.
Examples usage for each helper functions are given below and more detailed documentation can be found in the visualization tutorial.
[14]:
export_folder = 'exports'
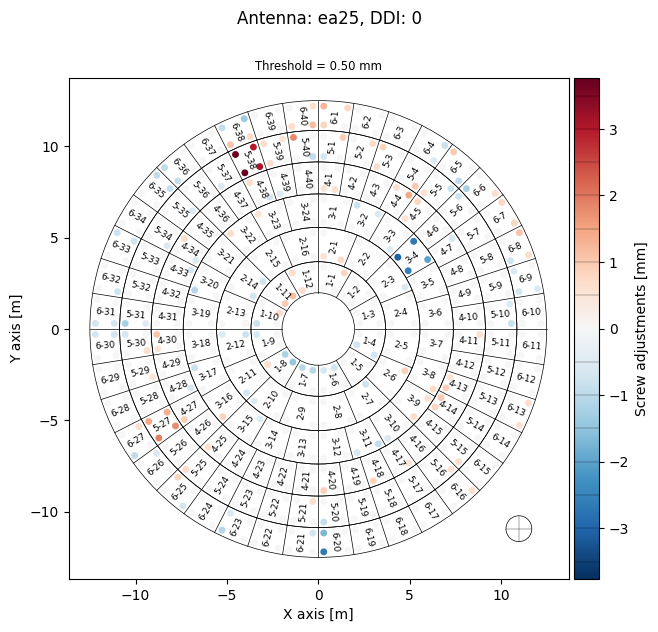
panel_mds.export_screws(
destination=export_folder,
ant='ea25',
ddi=0,
unit='mm',
threshold=0.5, # Threshold in mm for significant adjustments
display=True
)
[2024-02-14 12:53:43,938] INFO graphviper: Checking parameter values for mds.AstrohackPanelFile.export_screws
[2024-02-14 12:53:43,940] INFO astrohack: /export/home/ajax/jhoskins/Development/astrohack/
[2024-02-14 12:53:43,941] INFO graphviper: Searching /export/home/ajax/jhoskins/.conda/envs/viper-3.10/lib/python3.10/site-packages/graphviper/config/ for configuration file, please wait ...
[2024-02-14 12:53:43,946] INFO astrohack: Failed to find module in PARAMETER_CONFIG_PATH ... attempting to check common directories ...
[2024-02-14 12:53:43,948] INFO graphviper: Searching /export/home/ajax/jhoskins/Development/astrohack/ for configuration file, please wait ...
[2024-02-14 12:53:44,035] WARNING graphviper: Destination folder already exists, results may be overwritten
[15]:
with open(export_folder+'/panel_screws_ant_ea25_ddi_0.txt', 'r') as file:
for _ in range(30):
print(file.readline()[:-1])
# Screw adjustments for VLA ea25 antenna
# Adjustments are in mm
# Lower means away from subreflector
# Raise means toward the subreflector
# LOWER the panel if the number is POSITIVE
# RAISE the panel if the number is NEGATIVE
Panel il ir ol or
1-1 -0.15 0.21 0.15 0.83
1-2 -0.25 -0.36 0.35 0.13
1-3 0.41 0.39 0.01 -0.03
1-4 -0.43 -0.75 -0.09 -0.68
1-5 -0.39 -0.24 -0.40 -0.13
1-6 -0.74 -0.67 0.01 0.13
1-7 -1.04 -1.05 0.18 0.16
1-8 -1.45 -1.20 0.15 0.62
1-9 0.37 0.12 -0.14 -0.61
1-10 0.32 0.71 -0.84 -0.11
1-11 0.95 1.16 -0.37 0.01
1-12 0.63 0.11 0.70 -0.27
2-1 0.57 0.74 0.24 0.49
2-2 -0.26 -0.35 0.03 -0.10
2-3 0.29 0.20 -0.43 -0.56
2-4 -0.08 -0.08 -0.18 -0.18
2-5 -0.49 0.10 -0.45 0.45
2-6 0.27 -0.31 0.89 0.01
2-7 -0.80 -0.50 -0.40 0.06
2-8 0.22 0.11 -0.24 -0.42
[16]:
panel_mds.plot_antennas(
destination=export_folder,
ant='ea25',
ddi=0,
plot_type='deviation',
plot_screws=False,
dpi=300,
parallel=False,
display=True
)
[2024-02-14 12:53:47,289] INFO graphviper: Checking parameter values for mds.AstrohackPanelFile.plot_antennas
[2024-02-14 12:53:47,292] INFO astrohack: /export/home/ajax/jhoskins/Development/astrohack/
[2024-02-14 12:53:47,293] INFO graphviper: Searching /export/home/ajax/jhoskins/.conda/envs/viper-3.10/lib/python3.10/site-packages/graphviper/config/ for configuration file, please wait ...
[2024-02-14 12:53:47,294] INFO astrohack: Failed to find module in PARAMETER_CONFIG_PATH ... attempting to check common directories ...
[2024-02-14 12:53:47,298] INFO graphviper: Searching /export/home/ajax/jhoskins/Development/astrohack/ for configuration file, please wait ...
[2024-02-14 12:53:47,365] WARNING graphviper: Destination folder already exists, results may be overwritten